library(cmdstanr)
library(ggplot2)
library(tidyverse)
library(tidybayes)
library(targets)tar_load(one_time)
plot(one_time, type = "l")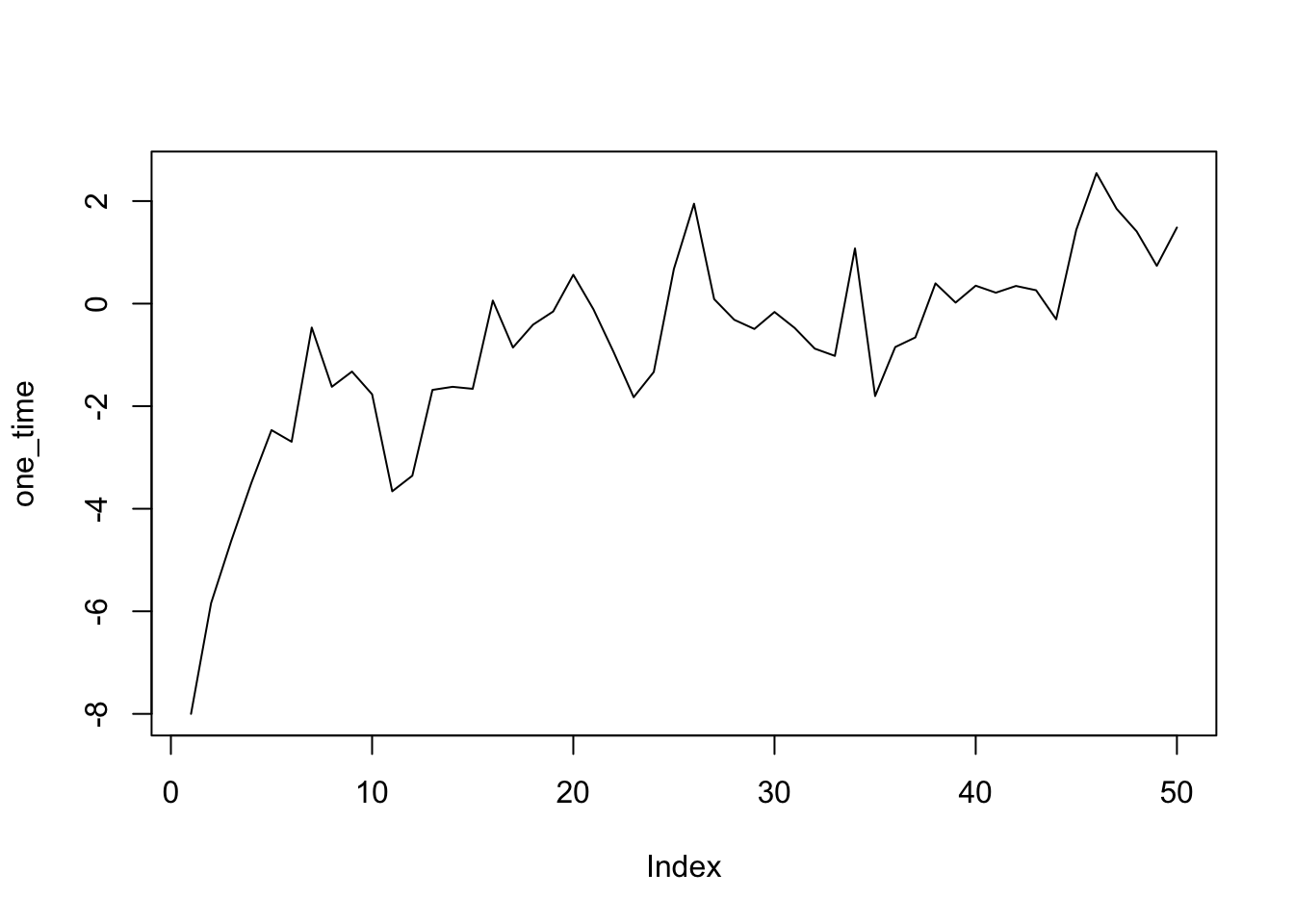
Controversy!
# load the model in stan
transition <- cmdstan_model(here::here("posts/2023-11-24-how-to-model-growth/ar1.stan"))
transitiondata{
int n;
vector[n] time;
vector[n] x;
}
// transformed data {
// vector[n] x = log(pop);
// }
parameters {
real<lower=0> a;
real<lower=0,upper=1> b;
real<lower=0> sigma;
}
transformed parameters {
real mu_max = a / (1 - b);
real sigma_max = sigma /sqrt(1 - b^2);
}
model {
a ~ normal(2, .5);
b ~ beta(5,2);
sigma ~ exponential(5);
x ~ normal(
mu_max .* (1 - pow(b, time)),
sigma_max .* sqrt(1 - pow(b^2, time))
);
}
generated quantities {
vector[15] x_pred;
x_pred[1] = 0;
for (j in 1:14) {
x_pred[j+1] = normal_rng(
mu_max * (1 - pow(b, j)),
sigma_max * sqrt(1 - pow(b^2, j))
);
}
}Here is another approach, using a lagged population growth model
# load the model in stan
lagged_growth <- cmdstan_model(here::here("posts/2023-11-24-how-to-model-growth/multiple_spp_ar1.stan"))
lagged_growthdata {
int n;
int nclone;
vector[n] x;
array[n] int<lower=1, upper=nclone> clone_id;
// for predictions
int<lower=0, upper=1> fit;
int nyear;
}
transformed data {
array[n - nclone] int time;
array[n - nclone] int time_m1;
for (i in 2:n) {
if (clone_id[i] == clone_id[i-1]) {
time[i - clone_id[i]] = i;
time_m1[i - clone_id[i]] = i - 1;
}
}
}
parameters {
real<lower=0> a;
real<lower=0,upper=1> b;
real<lower=0> sigma;
}
model {
a ~ normal(2, .5);
b ~ beta(5,2);
sigma ~ exponential(5);
if (fit == 1) {
x[time] ~ normal(
a + b * x[time_m1],
sigma);
}
}
generated quantities {
vector[nyear] x_pred;
x_pred[1] = 0;
for (j in 2:nyear){
x_pred[j] = a + b * x_pred[j-1] + normal_rng(0, sigma);
}
}Simulations
Here are simulations from a one-species AR-1 model that imitate Ives et al. figure 1.
simulate_pop_growth <- function(
a = 0,
b,
sigma = 1,
tmax = 50,
x0 = -8) {
xvec <- numeric(tmax)
xvec[1] <- x0
## process error
eta <- rnorm(tmax, mean = 0, sd = sigma)
for(time in 2:tmax){
xvec[time] <- a + b*xvec[time-1] + eta[time]
}
return(xvec)
}I’m going to simulate a modest number of time series, and choose parameters to make the time series slightly resemble the aphid experiment.
a_fig = 1
b_fig = .8
sigma_fig = .7
ts_data <- map_dfr(1:12,
~ tibble(
pop = simulate_pop_growth(
a = a_fig,
b = b_fig,
tmax = 16,
sigma = sigma_fig,
x0 = 0
),
time = 0:(length(pop)-1)
),
.id = "sim"
)
ts_data |>
ggplot(aes(x =time, y = pop, group = sim)) +
geom_line()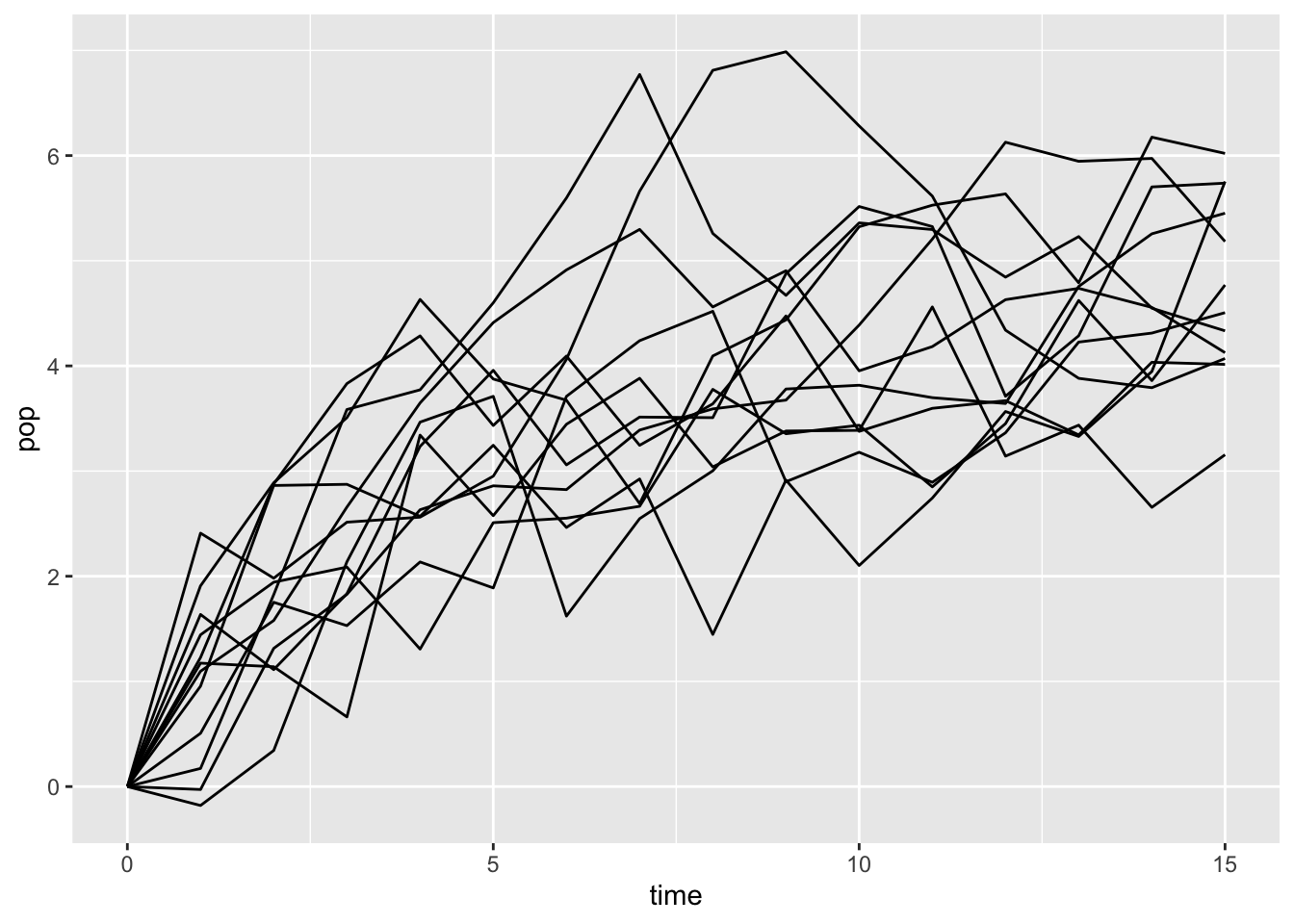
knitr::kable(head(ts_data))| sim | pop | time |
|---|---|---|
| 1 | 0.0000000 | 0 |
| 1 | 0.9548568 | 1 |
| 1 | 2.8629644 | 2 |
| 1 | 2.8740721 | 3 |
| 1 | 2.5695696 | 4 |
| 1 | 3.2455612 | 5 |
Transition distribution
ts_data_nozero <- filter(ts_data, time != 0)
transition_sample <- transition$sample(
data = list(n = nrow(ts_data_nozero),
x = ts_data_nozero$pop,
time = ts_data_nozero$time),
parallel_chains = 4, refresh = 0)Running MCMC with 4 parallel chains...
Chain 1 finished in 1.7 seconds.
Chain 3 finished in 1.7 seconds.
Chain 2 finished in 1.8 seconds.
Chain 4 finished in 1.7 seconds.
All 4 chains finished successfully.
Mean chain execution time: 1.7 seconds.
Total execution time: 2.0 seconds.transition_sample |>
spread_rvars(x_pred[time]) |>
ggplot(aes(x = time-1, ydist = x_pred)) +
stat_lineribbon() +
scale_fill_brewer(palette = "Greens", direction = -1) +
theme_bw() +
geom_line(aes(x = time, y = pop, group = sim),
inherit.aes = FALSE, data = ts_data) +
labs(x = "Time", y = "log population size")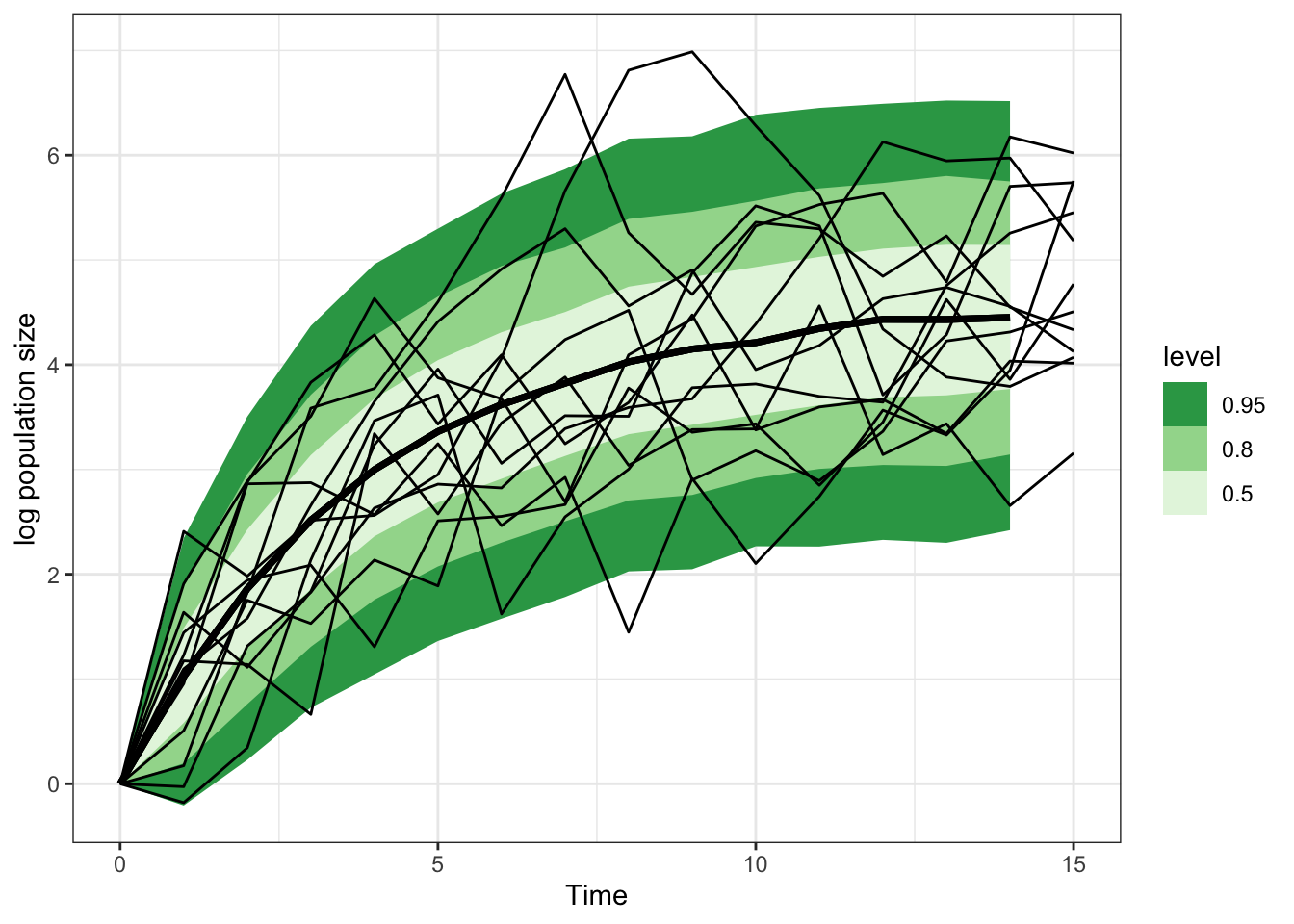
transition_sample |>
gather_rvars(a, b, sigma) |>
ggplot(aes(y = .variable, dist = .value)) +
stat_halfeye() +
geom_point(
aes(y = .variable, x = .value),
inherit.aes = FALSE,
data = tribble(
~ .variable, ~.value,
"a", a_fig,
"b", b_fig,
"sigma", sigma_fig), col = "red", size = 2) 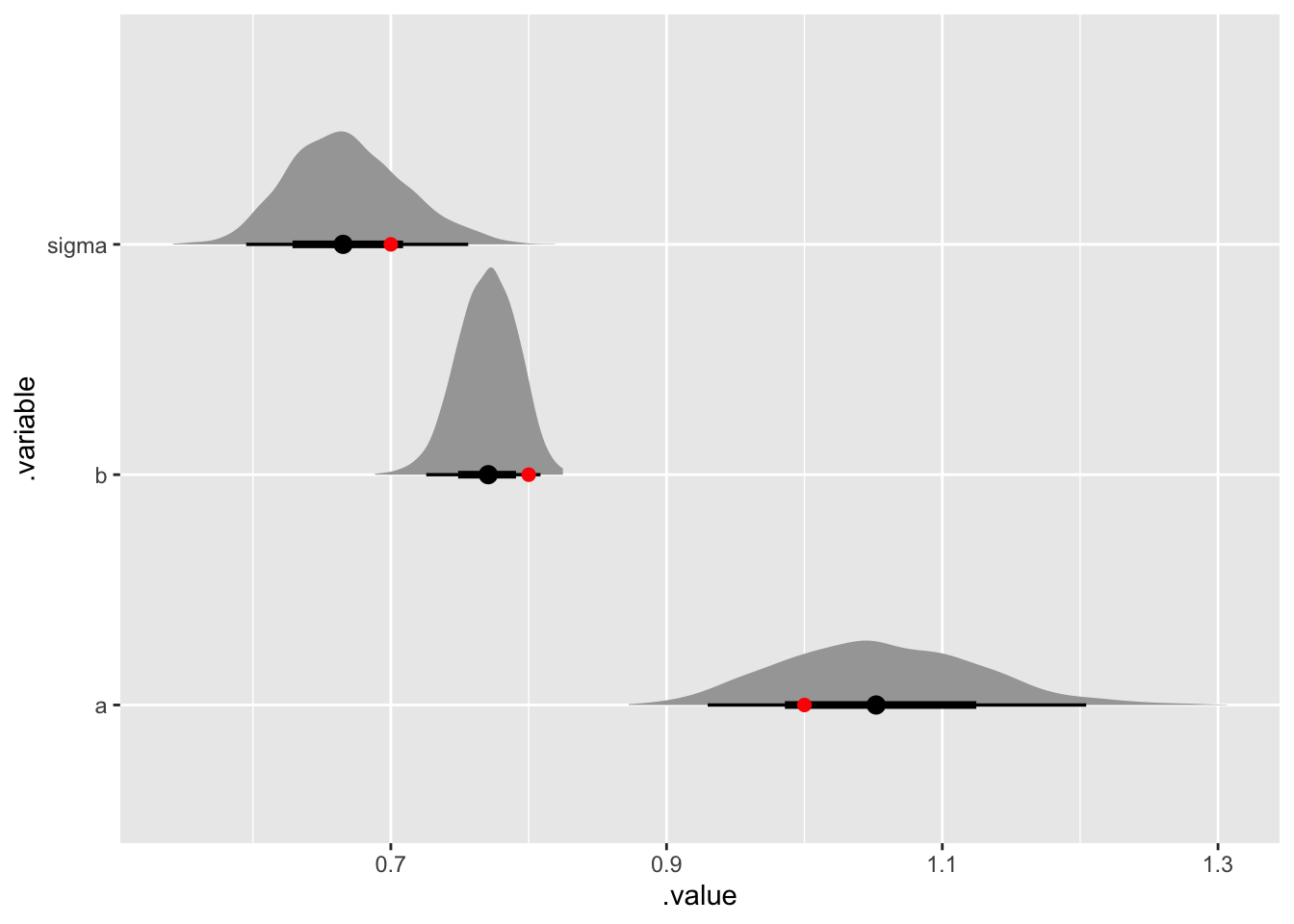
Lagged model
This time there is no need to drop 0s
ts_data <- ts_data |>
mutate(sim = readr::parse_number(sim))
lagged_growth_sample <- lagged_growth$sample(
data = list(n = nrow(ts_data),
nclone = max(ts_data$sim),
x = ts_data$pop,
time = ts_data$time,
clone_id = ts_data$sim,
fit = 1,
nyear = 15),
parallel_chains = 4, refresh = 0)Running MCMC with 4 parallel chains...
Chain 1 finished in 0.3 seconds.
Chain 2 finished in 0.3 seconds.
Chain 3 finished in 0.3 seconds.
Chain 4 finished in 0.3 seconds.
All 4 chains finished successfully.
Mean chain execution time: 0.3 seconds.
Total execution time: 0.5 seconds.predictions
lagged_growth_sample |>
spread_rvars(x_pred[time]) |>
ggplot(aes(x = time-1, ydist = x_pred)) +
stat_lineribbon() +
scale_fill_brewer(palette = "Greens", direction = -1) +
theme_bw() +
geom_line(aes(x = time, y = pop, group = sim),
inherit.aes = FALSE, data = ts_data) +
labs(x = "Time", y = "log population size")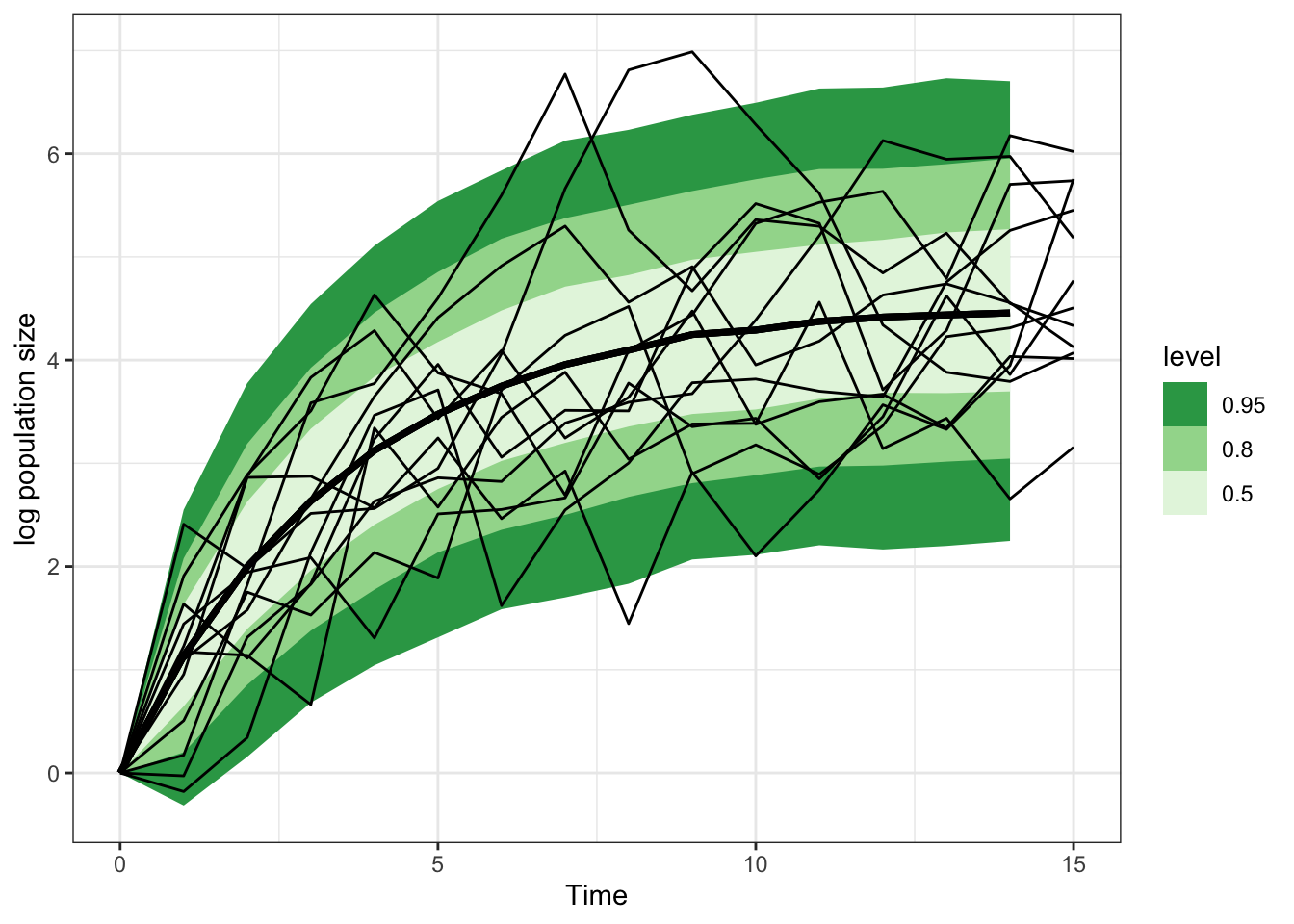
parameters
lagged_growth_sample |>
gather_rvars(a, b, sigma) |>
ggplot(aes(y = .variable, dist = .value)) +
stat_halfeye() +
geom_point(
aes(y = .variable, x = .value),
inherit.aes = FALSE,
data = tribble(
~ .variable, ~.value,
"a", a_fig,
"b", b_fig,
"sigma", sigma_fig), col = "red", size = 2) 