library(targets)
library(ggplot2)
library(tidyverse)
library(tidybayes)You can give uncorrelated random numbers a specific correlation by using the Cholesky decomposition. This comes in handy when you’re modelling correlated random variables using MCMC.
eg <- rethinking::rlkjcorr(1, 2, 1)
eg [,1] [,2]
[1,] 1.0000000 -0.5587252
[2,] -0.5587252 1.0000000cc <- chol(eg)
purrr::rerun(30,{
zz <- matrix(data = rnorm(500), ncol = 2)
# plot(zz)
rr <- t(cc) %*% t(zz)
# plot(t(rr))
cor(t(rr))[1,2]
}) |>
purrr::flatten_dbl() |> density() |> plot(main = "Simulated correlations")Warning: `rerun()` was deprecated in purrr 1.0.0.
ℹ Please use `map()` instead.
# Previously
rerun(30, {
zz <- matrix(data = rnorm(500), ncol = 2)
rr <- t(cc) %*% t(zz)
cor(t(rr))[1, 2]
})
# Now
map(1:30, ~ {
zz <- matrix(data = rnorm(500), ncol = 2)
rr <- t(cc) %*% t(zz)
cor(t(rr))[1, 2]
})abline(v=eg[1,2])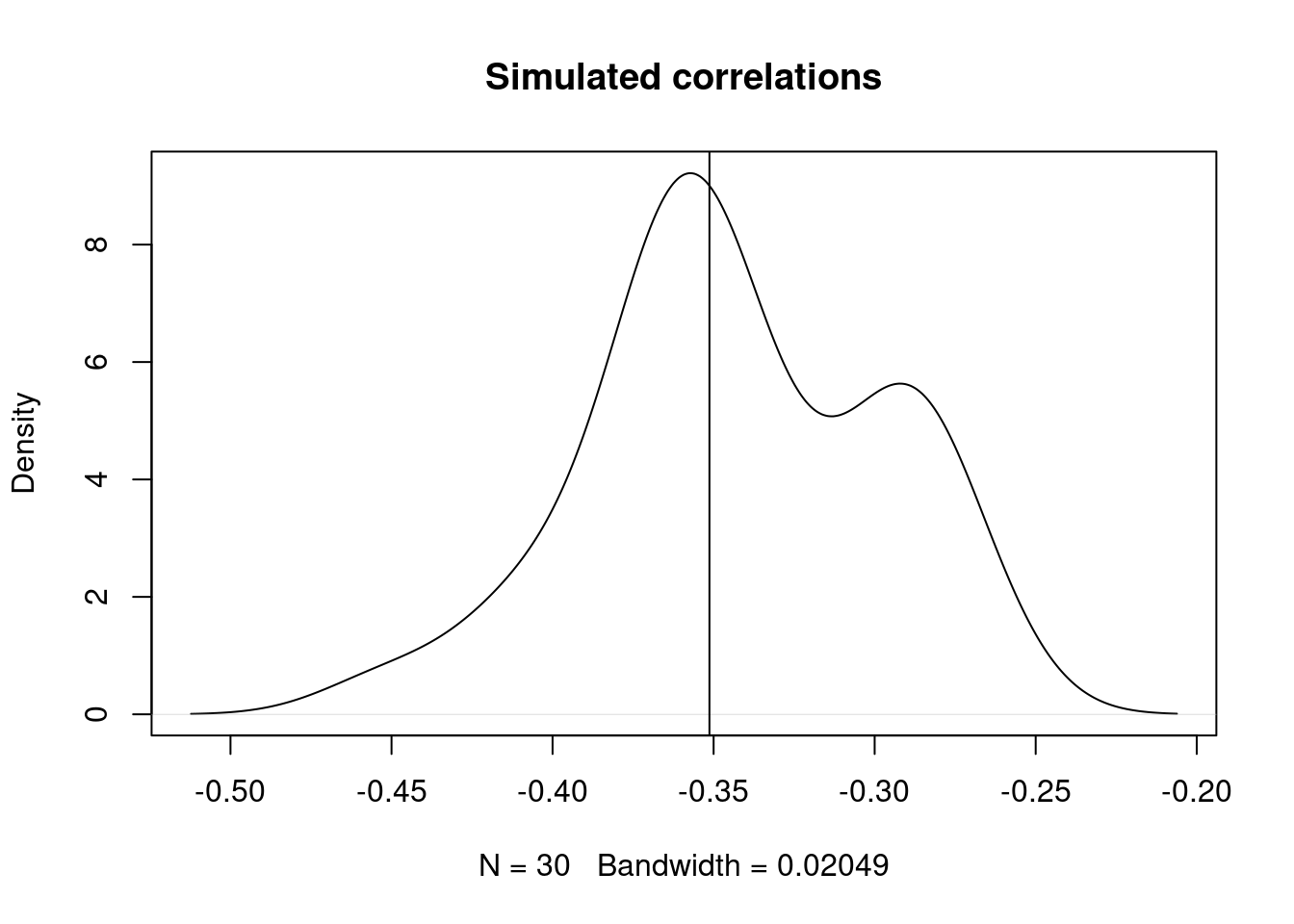
Here we can see that the correlation we want is -0.5587252, and indeed we’re able to give exactly that correlation to uncorrelated random numbers.
eg <- rethinking::rlkjcorr(1, 2, 5)
print(eg)
cc <- chol(eg)
t(cc) %*% cc
mm <- matrix(rnorm(4000, mean = 0, sd = 1), ncol = 2)
plot(mm)
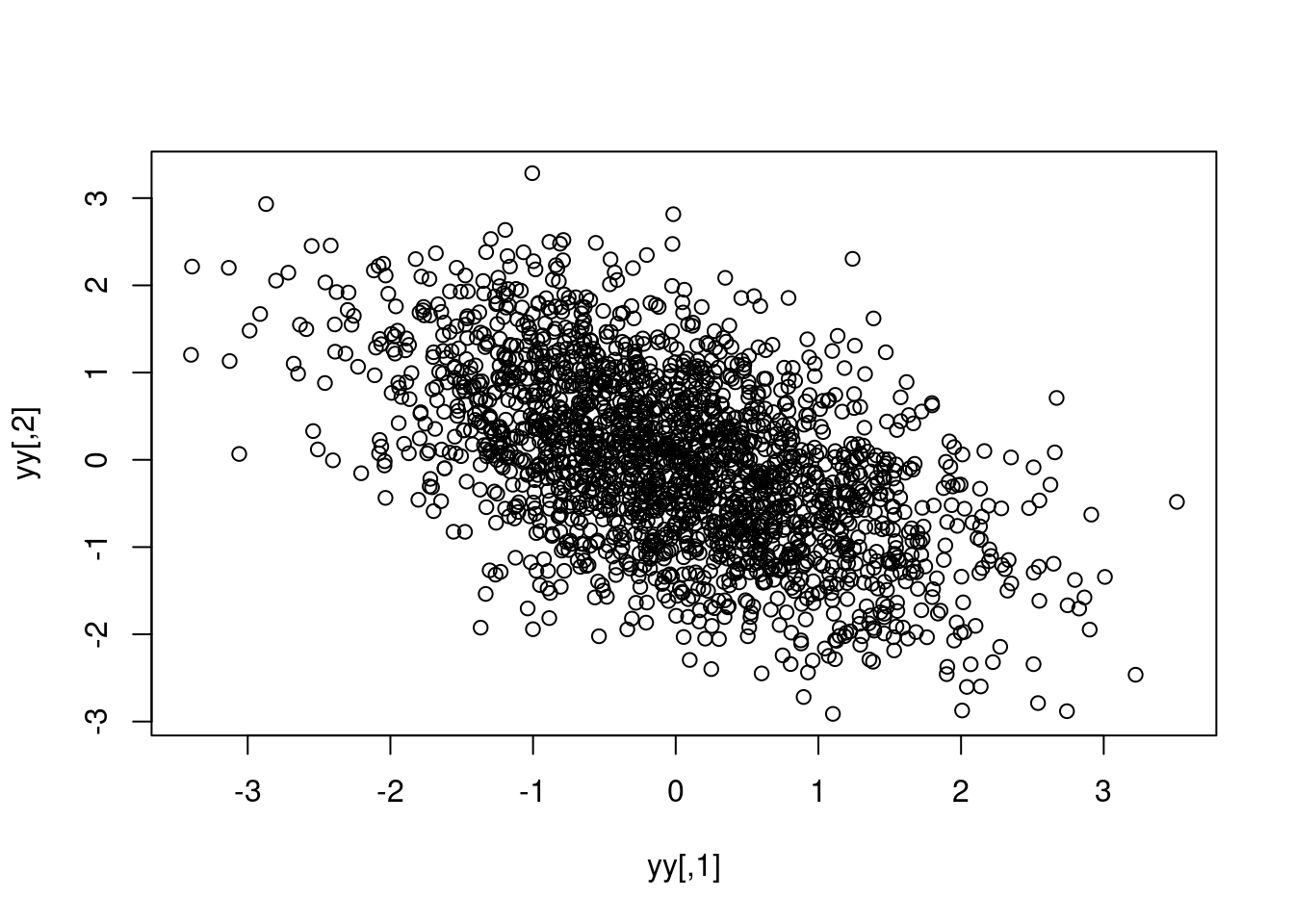
yy <- t(t(cc) %*% t(mm))
plot(yy) [,1] [,2]
[1,] 1.00000000 0.08358276
[2,] 0.08358276 1.00000000 [,1] [,2]
[1,] 1.00000000 0.08358276
[2,] 0.08358276 1.00000000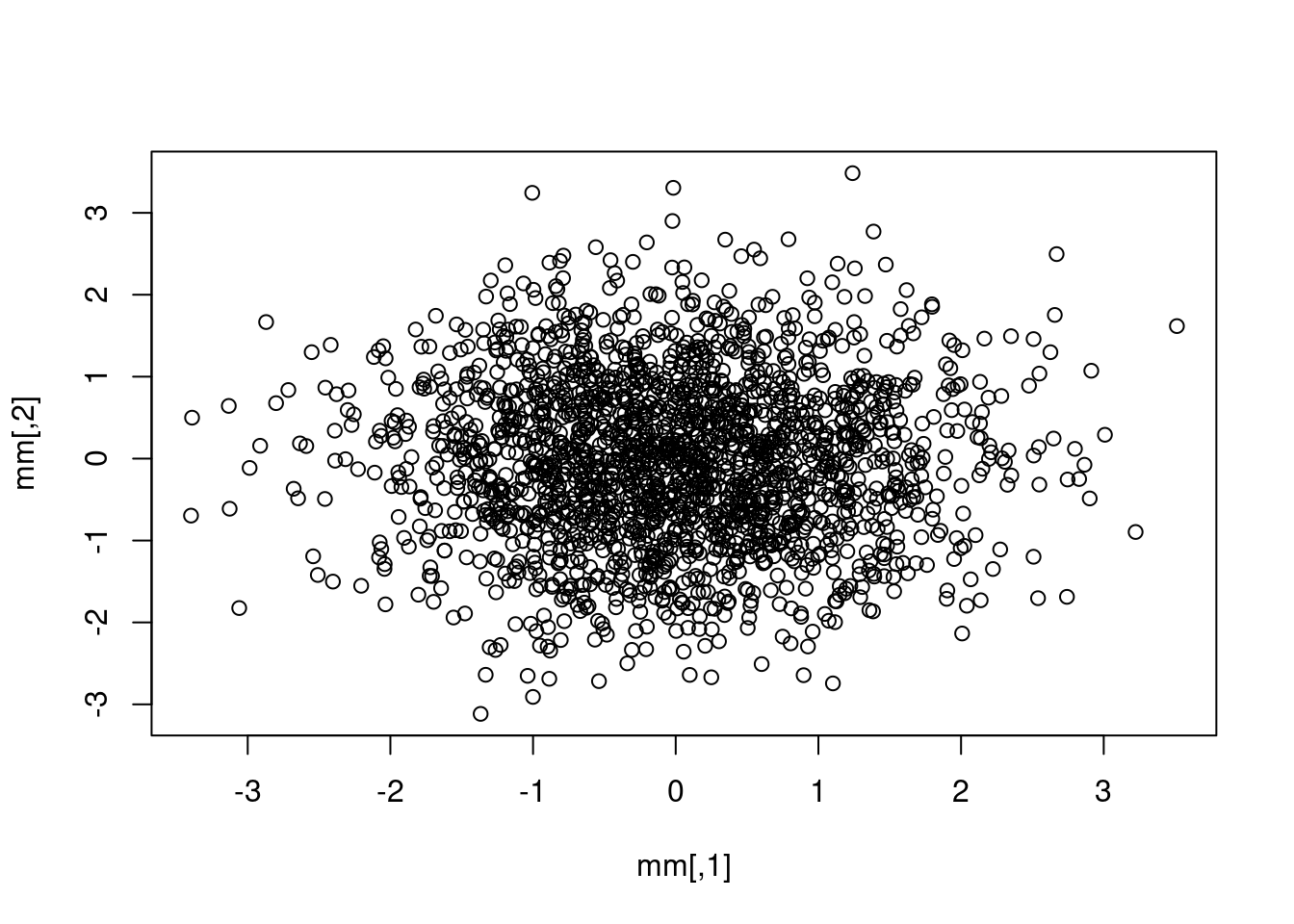
Doing it by hand
Because the case for only two random variables is pretty simple, we can actually write out the Cholesky decomposition by hand. Here I want to give some independent random numbers a correlation of -0.8:
p <- -.8
L <- matrix(c(1,0, p, sqrt(1 - p^2)), ncol = 2, byrow = TRUE)
zz <- matrix(data = rnorm(1000), ncol = 2)
yy <- t(L %*% t(zz))
plot(yy)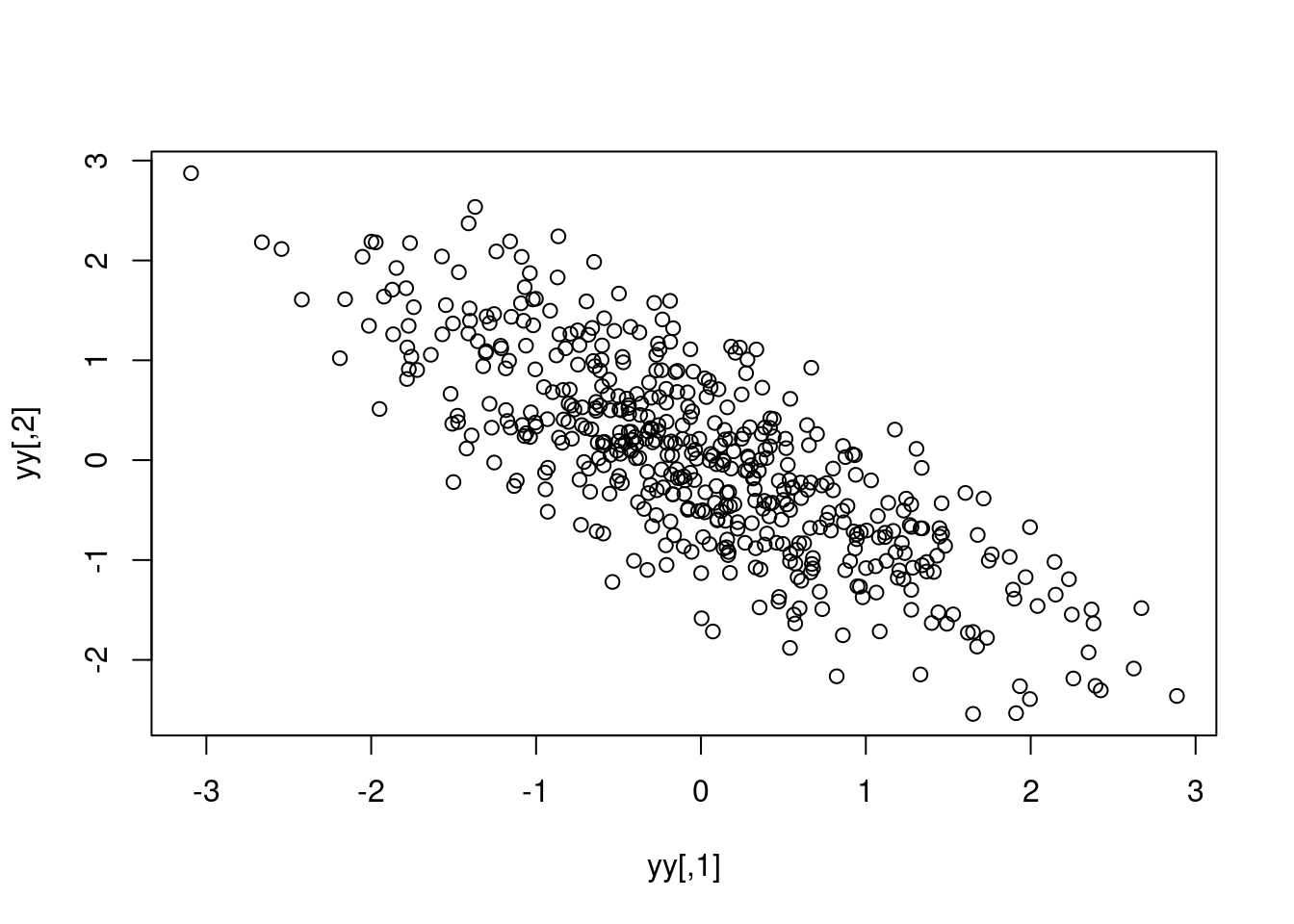
cor(yy) [,1] [,2]
[1,] 1.0000000 -0.8005567
[2,] -0.8005567 1.0000000This can be written without matrix multiplication like this:
z1 <- rnorm(1000)
z2 <- rnorm(1000)
y1 <- z1
y2 <- p*z1 + sqrt(1 - p^2)*z2
plot(y1, y2)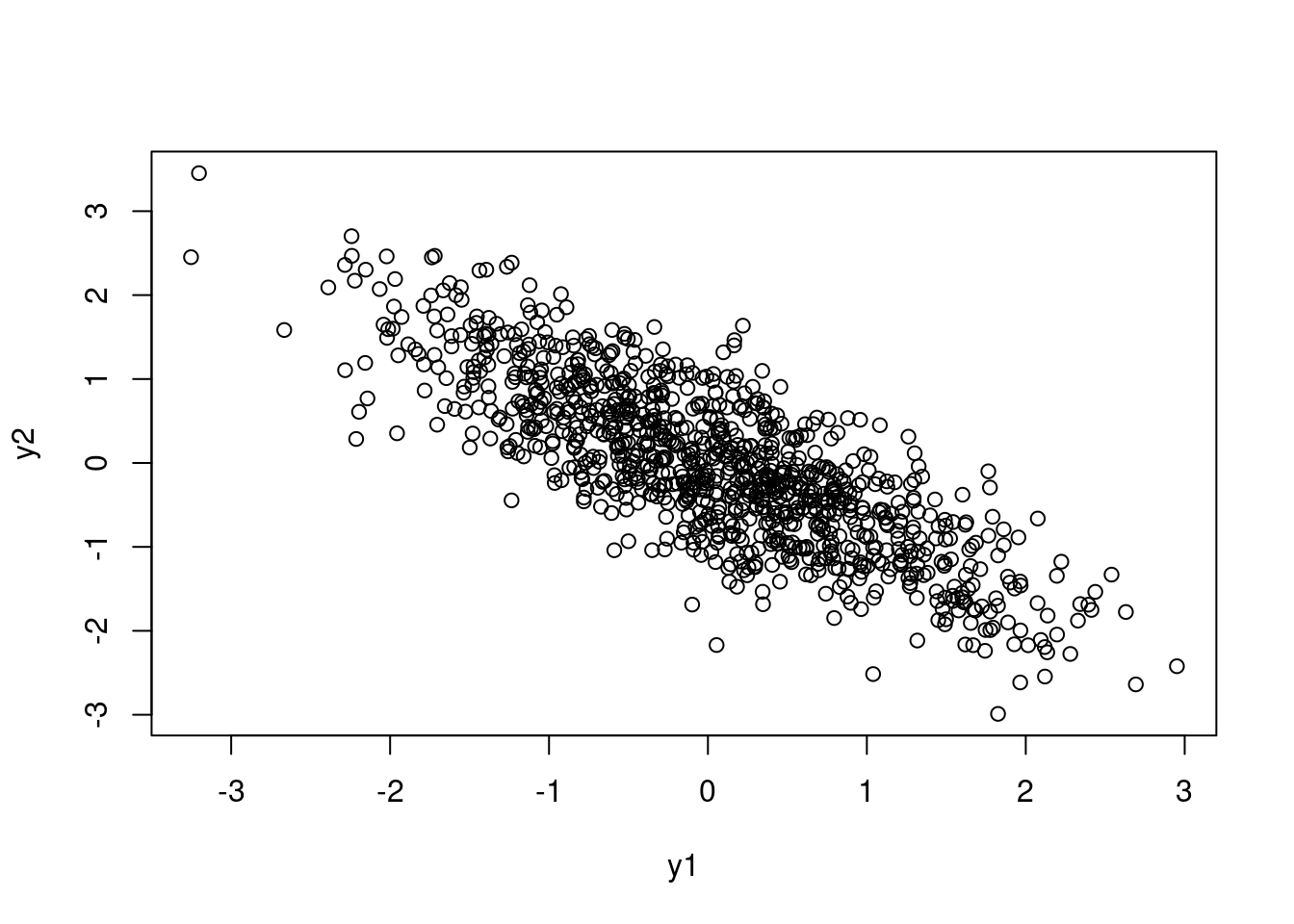
cor(y2, y1)[1] -0.8105853In a Stan model, I might want to use a scaled beta distribution to model the correlation between, say, slopes and intercepts in a model:
mu <- .84
phi <- 80
g <- rbeta(1, mu*phi, (1 - mu)*phi)
p_trans <- g*2 - 1
sim_y_corr <- function(n, p) {
z1 <- rnorm(n)
z2 <- rnorm(n)
y1 <- z1
y2 <- p*z1 + sqrt(1 - p^2)*z2
return(data.frame(y1, y2))
}
sim_y_corr(n = 1000, p = p_trans) |> plot()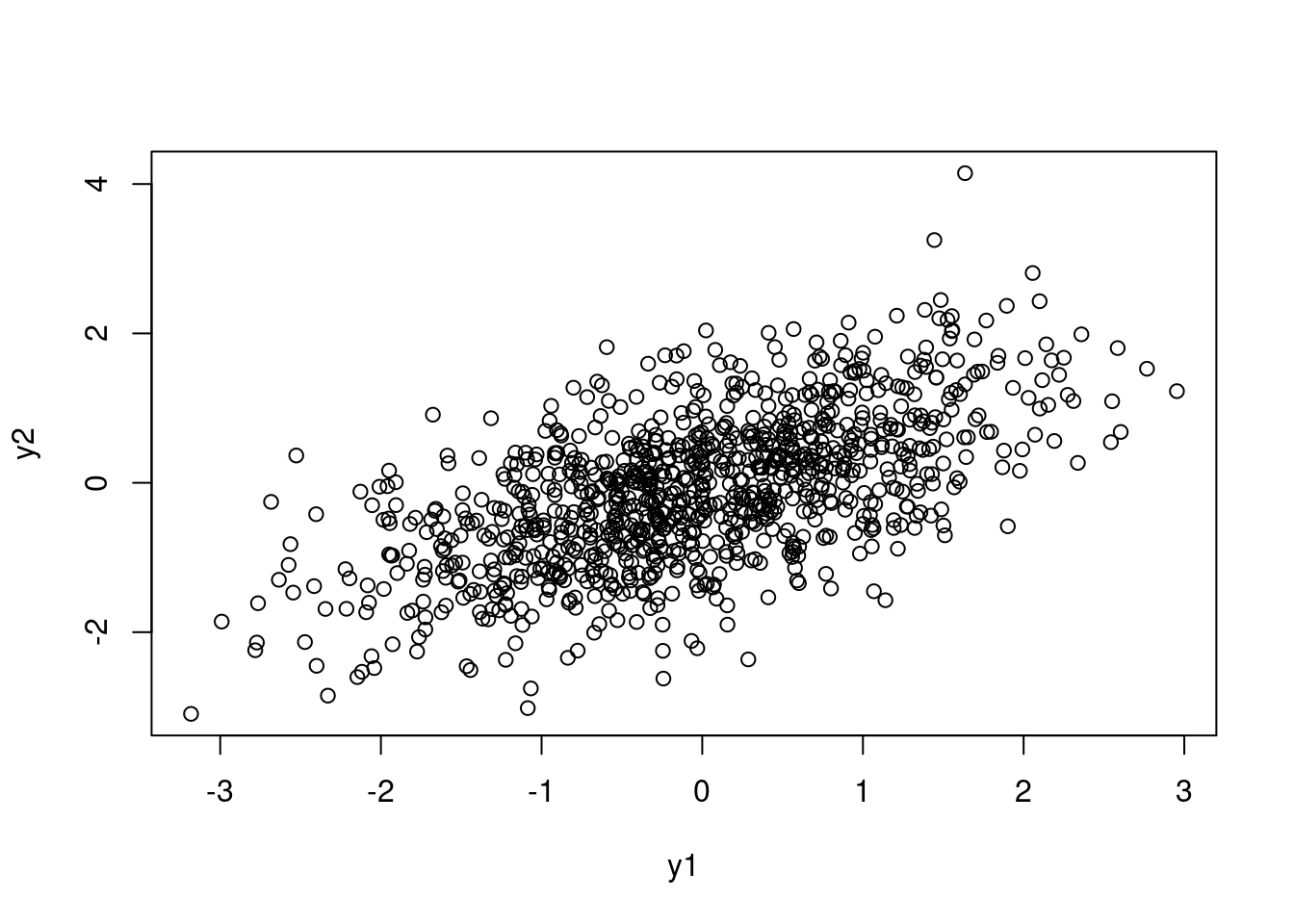
or even work on the logit scale:
q <- rnorm(1, mean = -1, sd = .1)
p_trans_n <- plogis(q)*2 - 1
sim_y_corr(n = 1000, p = p_trans_n) |> plot()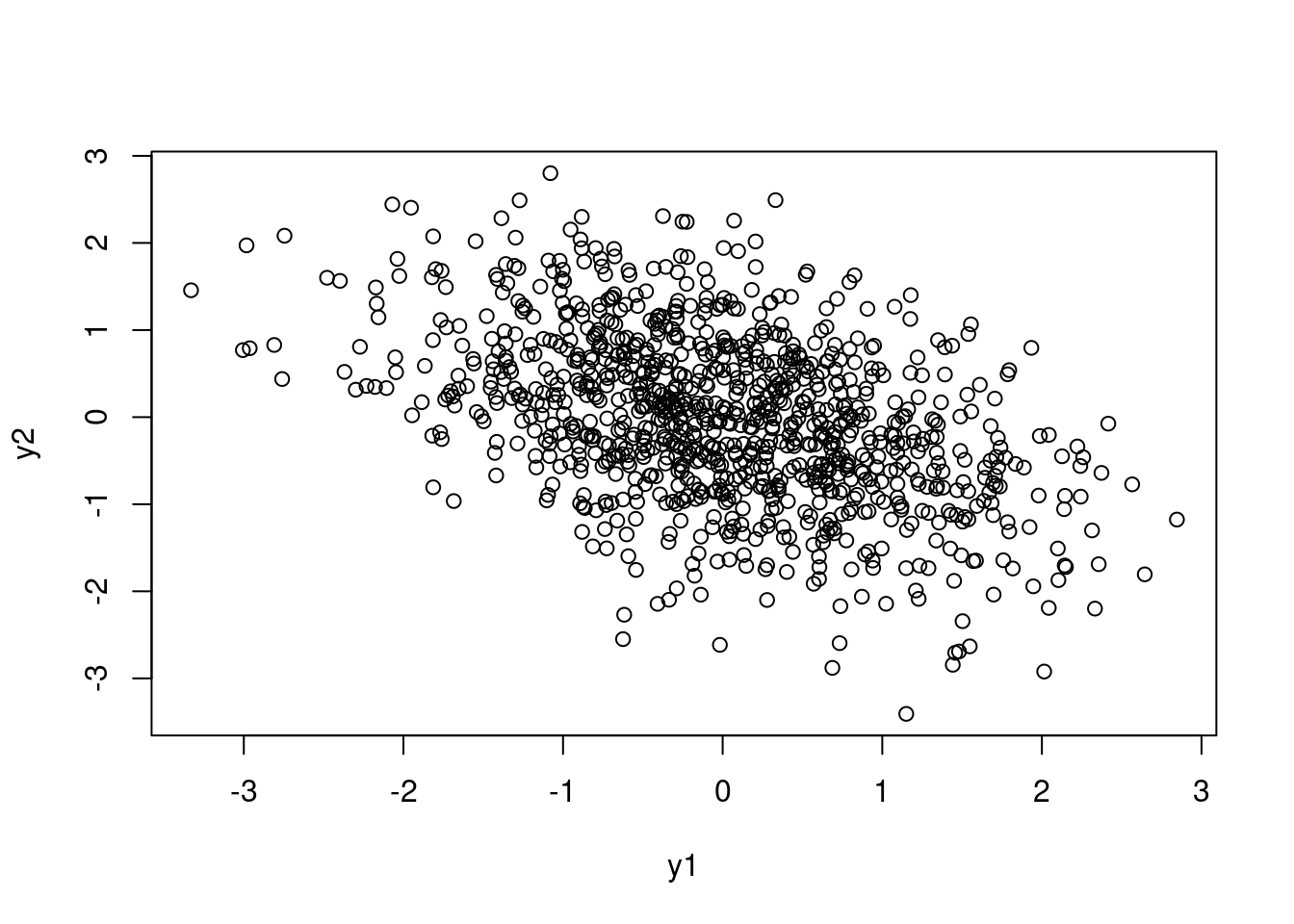
This is even a little easier to think about: because of all the transformations, 0 on the logit scale still means a 0 correlation; negative and positive numbers mean negative and positive correlations as well.
Another way to make correlation matrices is here: https://www.rdatagen.net/post/2023-02-14-flexible-correlation-generation-an-update-to-gencorgen-in-simstudy/